Euglenida
euglenids or euglenoids
Brian S. Leander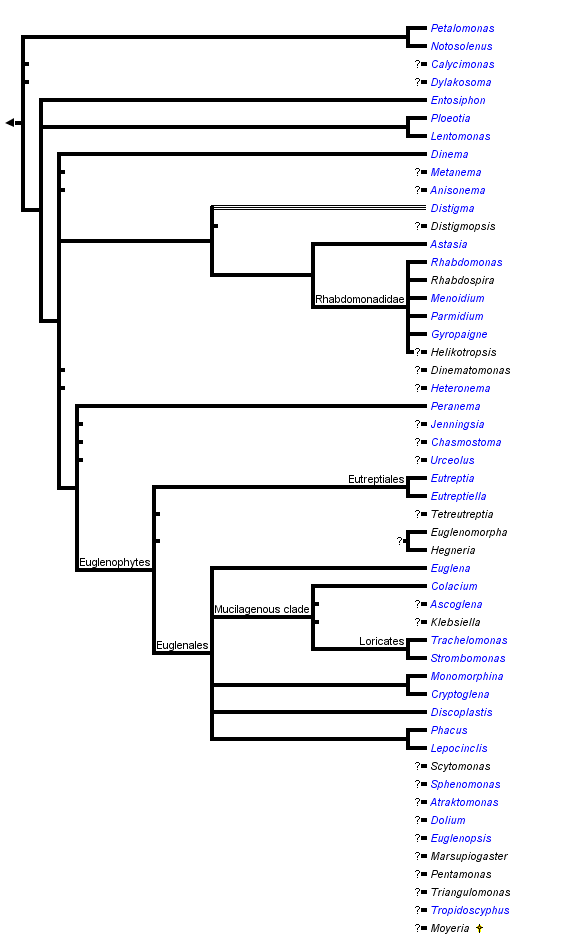

This tree diagram shows the relationships between several groups of organisms.
The root of the current tree connects the organisms featured in this tree to their containing group and the rest of the Tree of Life. The basal branching point in the tree represents the ancestor of the other groups in the tree. This ancestor diversified over time into several descendent subgroups, which are represented as internal nodes and terminal taxa to the right.

You can click on the root to travel down the Tree of Life all the way to the root of all Life, and you can click on the names of descendent subgroups to travel up the Tree of Life all the way to individual species.
For more information on ToL tree formatting, please see Interpreting the Tree or Classification. To learn more about phylogenetic trees, please visit our Phylogenetic Biology pages.
close boxIntroduction
Euglenids (sometimes referred to as “euglenoids”) are a prominent group of free-living, aquatic flagellates with diverse modes of nutrition, including bacterivory (generally consumers of small prey cells like bacteria), eukaryovory (generally consumers of large prey cells like other microeukaryotes), osmotrophy (absorbers of organic molecules) and photoautotrophy (referred to as "euglenophytes"). The presence of both phagotrophic and photautotrophic species within the same group has led to redundant classification schemes under the International Code of Botanical Nomenclature and the International Code of Zoological Nomenclature (so-called “ambiregnal taxa”). Euglenids are neither plant nor animal, so the group does not fall neatly within the archaic plant-animal dichotomy that still pervades biological thinking.
Approximately 1,400 species of euglenids have been described so far, and it is possible that at least twice that many await discovery. The amount of morphological and behavioral diversity present in this group is exceptionally high (e.g. see title illustrations and Fig. 1) and provides compelling evidence for major events in eukaryote evolution, such as the punctuated effects of secondary endosymbiosis and mutations in underlying developmental mechanisms (Leander et al. 2007). Several photosynthetic and osmotrophic species are bloom-formers in nutrient-rich conditions and are useful indicators of environmental pollution. Phagotrophic species (i.e. bacterivores and eukaryovores) are ubiquitous primary consumers and are important components of microbial food webs across the globe. Knowledge of euglenids extends back to the invention of the first microscope in the late 1600’s by Leeuwenhoek, and a few of these species have subsequently been used as model systems for addressing a wide variety of questions in basic cell biology. Euglena gracilis, for instance, is familiar to nearly every student who has ever taken a general biology course in high school, college or university.

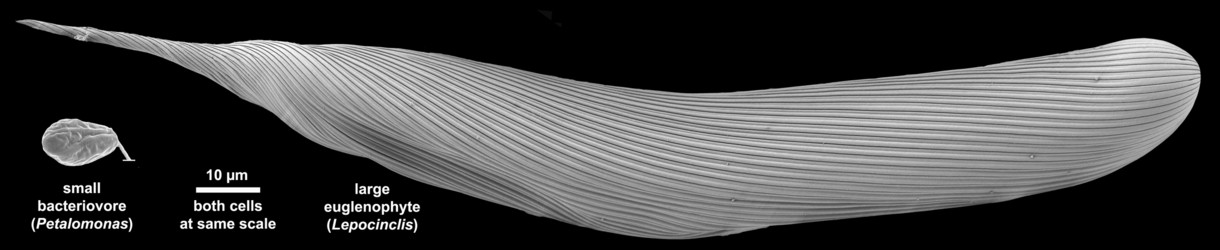
Fig. 1. Scanning electron micrographs showing the relative number of pellicle strips and the size differences between a small bacterivore (left, with 4 pellicle strips) and a large euglenophyte (right, with 48 pellicle strips). © Brian S. Leander
Characteristics
- Mitochondria with discoidal (paddle-shaped) cristae.
- Conspicuous nucleus with permanently condensed chromosomes and nucleolus.
- Eukaryovorous, osmotrophic and photoautotrophic euglenids with crystallized carbohydrate storage products called paramylon (beta 1,3 glucan).
- Ejectile organelles in the form of tubular extrusomes, mucocysts, or muciferous bodies.
- Elaborate Golgi bodies.
- Usually two heterodynamic (gliding) flagella that emerge from an anterior flagellar pocket (Fig. 2A,B). Most photoautotropic and osmotrophic species have one emergent or swimming flagellum and one non-emergent flagellum that is reduced in length and confined to the flagellar pocket. Some species have two or more emergent flagella and some lack them altogether.
- Flagella with paraxial (paraxonemal) rods: the rod in the dorsal flagellum has a tubular construction and the rod in the ventral flagellum has a lattice-like construction.
- Flagellar apparatus consisting of a flagellar pocket, two basal bodies (dorsal and ventral) and three microtubular roots: the dorsal root, the ventral root and the intermediate root.
- The flagellar pocket in photoautotrophic species is modified into a reservoir and canal (Fig. 2E).
- Freshwater lineages have contractile vacuoles associated with the reservoir.
- Photoautotrophic species have chloroplasts surrounded by three membranes and with thylakoids in stacks of three. Pyrenoids present, except in Discoplastis, Phacus and Lepocinclis.
- Photoautotrophic species respond to the direction and intensity of light using a shading stigma (Fig. 2E,F) or eyespot and a photosensory swelling at the base of the emergent flagellum.
- Feeding apparatus consisting of a feeding pocket reinforced by microtubules from the ventral root. The feeding pocket in many bacteriovores and in eukaryovores is further reinforced by robust rods and vanes. The feeding pocket in photoautotrophic and osmotrophic species is highly reduced.
- Diverse and dynamic modes of locomotion, including metaboly, substrate-mediated gliding and swimming.
- Cytokinesis involves a longitudinal cleavage furrow.
- The best synapomorphy for the group is a complex pellicle consisting of S-shaped proteinaceous strips beneath the plasma membrane, superficial microtubules and cisternae of endoplasmic reticulum. The pellicle strips are oriented longitudinally in bacteriovorous euglenids (Fig. 2C) and helically in eukaryovorous, photoautotrophic and osmotrophic euglenids (Fig. 2D). The strips are secondarily longitudinal in some rigid photoautotrophs (e.g. Phacus) and osmotrophs (e.g. Menoidium).

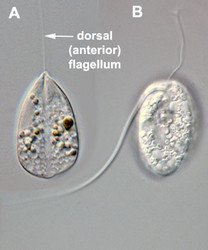
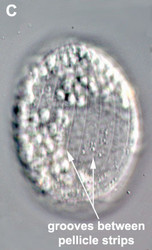
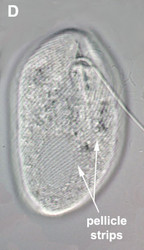
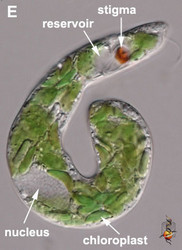
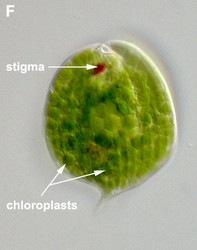
Fig. 2. Light micrographs showing general characteristics of euglenid cells. A-B. Two cells that glide along substrates showing a straight anterior flagellum (A) and a thick recurrent or posterior flagellum that trails the cell (B). C. A phagotrophic cell with about 20 relatively thick pellicle strips that are more or less longitudinally arranged. D. A phagotrophic cell with >40 thin pellicle strips that are helically arranged and capable of metaboly. E-F. Photoautotrophic cells showing chloroplasts, the nucleus, the reservoir and the stigma. Images A-D and F © Brian S. Leander. Image E © L. Amaral-Zettler and D. J. Patterson.
Habitat
Phagotrophic lineages are widespread in marine, brackish and freshwater sediments. These cells glide within the spaces between grains of sand and within the narrow interface between mud and the water column. Photoautotrophic and osmotrophic lineages mainly inhabit the water column of freshwater environments. However, relatively large and vermiform photoautotrophic species have reduced flagella and tend to creep within sediments. Several photoautotrophic species also inhabit brackish water and estuaries either in the sediments or by swimming in the water column. Only a few photoautotrophic lineages inhabit marine planktonic communities (e.g. the Eutreptiales). Some photoautotrophic species migrate vertically in marine sand in coordination with tidal and diurnal cycles (e.g. Euglena rustica and E. obtusa). These species are usually found in high abundance and form easily visible green patches in marine sand during low tides (Brown et al. 2002).
Photoautotrophic and osmotrophic euglenids can be found in particularly high abundance (blooms) in eutrophic freshwater environments and are indicators of organic pollution. Some bloom-forming (photoautotrophic) euglenids (e.g. Euglena sanguinea) have been shown to produce ichthyotoxins that cause fish die-offs (Zimba et al. 2004).
Flagellar Apparatus
Most euglenids possess two dynamic flagella that emerge from a flagellar pocket; however, a few lineages have more than two (e.g. Tetreutreptia has four flagella) and some have highly reduced flagella giving the appearance of one or none when viewed with the light microscope. The flagella of euglenids, and kinetoplastids, possess paraxial (paraxonemal) rods within the flagella that run side-by-side with the highly conserved 9-plus-2 microtubular axoneme found in most other eukaryotes. The paraxial rods make euglenid flagella conspicuously thick when viewed under the light microscope. The paraxial rod in each flagellum has a different ultrastructure: the paraxial rod in the ventral flagellum has a lattice-like structure, and the paraxial rod in the dorsal flagellum has a whorled structure. The flagellar membrane is adorned with hairs or mastigonemes.
In phagotrophic lineages, the flagella are heterodynamic, where one flagellum (i.e. the dorsal or anterior flagellum) is held straight in front of the cell and the other flagellum (i.e. the ventral, recurrent or posterior flagellum) is bent backwards, sits within a ventral groove or sulcus and trials the cell (see Fig. 3). The hairs and paraxial rods of these flagella facilitate gliding motility across substrates (Saito et al. 2003). The tip of the dorsal or anterior flagellum is in constant motion and functions as a sensory apparatus. Some phagotrophic euglenids use the anterior flagellum like a hook to shovel prey cells into the feeding apparatus (Breglia and Leander, unpubl. data). The flagella of euglenids are highly sensitive to mechanical stimulation and presumably respond to chemical and electrical stimuli as well; however, experiments in this regard have yet to be explored.
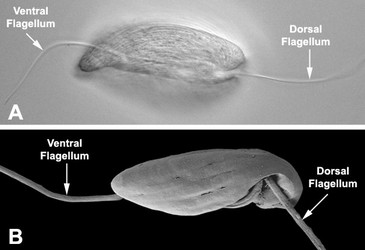
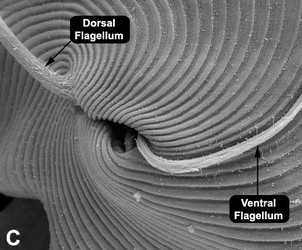
Fig. 3. Micrographs of the euglenid flagellar apparatus. A-B. Light micrograph (A) and scanning electron micrograph (B) showing the heterodynamic orientation of the dorsal (anterior) and ventral (posterior) flagella in gliding (phagotrophic) cells. C. Scanning electron micrograph of Peranema trichophorum showing two flagella emerging from the anterior opening of the flagellar pocket. © 2008 Brian S. Leander
Osmotrophic and photoautotrophic euglenids exploit the water column and have abandoned gliding motility for swimming motility (Leander 2004). Several primary osmotrophic euglenids (e.g. Distigma) and the Eutreptiales (photoautotrophs) possess two highly active flagella with erratic beat patterns. The majority of photoautotrophic and secondarily osmotrophic euglenids possess a reduced (non-emergent) flagellum and a dynamic flagellum that emerges from the canal. The emergent flagellum undergoes more organized and consistent beat patterns that take the form of a “figure-eight” or a lasso. This beat pattern functions much like an airplane propeller and pulls the euglenid cell through the water column.
The flagellar axonemes are anchored by basal bodies that are situated at the base of the flagellar pocket: the ventral flagellum emerges from the ventral basal body, and the dorsal flagellum emerges from the dorsal basal body. Three microtubular roots extend from the basal bodies: (1) a dorsal root extends from the lateral side of the dorsal basal body, (2) a ventral root extends from the lateral side of the ventral basal body, and (3) an intermediate root extends from the medial side of the ventral basal body and resides between both basal bodies. A striated fiber connects both basal bodies. This flagellar apparatus constitutes the organizing center from which all other cytoskeletal elements of the euglenid cell originate – such as the microtubules associated with the cell surface (or pellicle) and the feeding apparatus (Belhadri et al. 1992; Belhadri and Brugerolle 1992; Farmer and Triemer 1988; Hilenski and Walne 1985; Leander 2004; Shin et al. 2001; Shin et al. 2002; Simpson 1997; Solomon et al. 1987; Surek and Melkonian 1986; Willey and Wibel 1985).
Pellicle and Metaboly
The best synapomorphy for the Euglenida is a novel cytoskeleton comprised of parallel proteinaceous strips and microtubules (Leander 2001, 2004; Leander and Farmer 2000a; Leander and Farmer 2001a; Leander et al. 2001). These elements are positioned beneath the plasma membrane and are closely associated with tubular cisternae of endoplasmic reticulum. Collectively, this ultrastructural system is referred to as the euglenid pellicle (Fig. 4).


Fig. 4. Organization and main components of the euglenid pellicle. © 2007 Heather Esson & Brian Leander (Modified from Leander et al. 2007)
The strips are composed mostly of a family of proteins called “articulins” (Marrs and Bouck 1992). The strips run along the length of the cell and may be arranged longitudinally or helically, depending on the species. In general, the main frame of each pellicle strip is “S-shaped” in cross section and consists of an arch region and a heel region that defines a groove (Leander et al. 2007; Leander and Farmer 2001a, Fig. 4). Adjacent strips articulate along their lateral margins; the strip arch overlaps with the heel of a neighboring strip, giving the surface of euglenid cells a striated appearance. The articulation zones between adjacent strips are discontinuities in the cell surface that facilitate (1) dynamic changes in cell shape, called metaboly or euglenoid movement (see Fig. 5; also see movies at The Euglenoid Project), and (2) cytoskeletal replication prior to cell division (i.e. cytokinesis). Metaboly is correlated with cells that have a large number of pellicle strips (over 20) and is thought to facilitate the ingestion of large food particles, such as other eukaryotic cells (Leander 2004; Leander et al. 2001, 2007). Metaboly also corresponds to the origin of eukaryovory in euglenids and set the stage for the secondary endosymbiotic event that led to the origin of photosynthesis in a diverse subgroup of euglenids (Leander 2004; Leander et al. 2001). Accordingly, many early diverging lineages of primay osmotrophic and photoautotrophic euglenids are still capable of metaboly (Fig. 5); this feature is a vestige of their eukaryovorous ancestry.


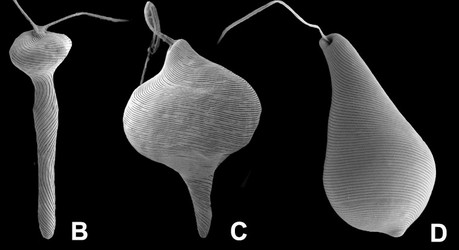
Fig. 5. Micrographs illustrating metaboly or euglenoid movement. A. Light micrographs showing three different stages of the peristalsis-like movements of Astasia (sensu stricto), a primary osmotrophic euglenid related to Distigma. B-D. Scanning electron micrographs showing metaboly in different euglenids: (B) Distigma, (C) Eutreptia and (D) Euglena. © 2004 A: William Bourland; B-D: Brian Leander.
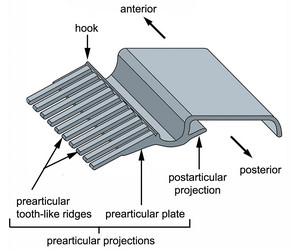
Fig. 6. Isolated proteinaceous strip of the euglenid pellicle illustrating the complexity of the prearticular projections in many photoautotrophic euglenids. © 2007 Heather Esson & Brian Leander (Modified from Leander et al. 2007)
In photoautotrophic and secondary osmotrophic lineages, the frame of each strip contains periodic arrays of projections that branch laterally from the heel (Leander et al. 2001, 2007; Leander and Farmer 2001a, Fig. 6). The projections of one strip articulate with the projections of an adjacent strip beneath the arches. The projections that branch beneath the arch of an adjacent strip, so-called “prearticular projections”, and those that branch beneath the arch of the same strip, so-called “postarticular projections”, can vary considerably in robustness: some lineages possess delicate thread-like projections; some species possess more robust tooth-like projections; and some species possess projections that form robust continuous plates. Euglenid cells with more delicate strips tend to demonstrate more dramatic degrees of metaboly; euglenid cells with robust strips tend to be rigid, or nearly so. Phagotrophic and primary osmotrophic euglenids lack strip projections altogether, which is an ancestral state for the Euglenida as a whole (Angeler et al. 1999; Leander 2004; Leander et al. 2001, 2007; Leander and Farmer 2001a).
The euglenid pellicle is multigenerational; each strip or cohort of strips represents a different cytokinetic event in the history of any particular cell (Esson and Leander 2006, 2008). Prior to cytokinesis, the number of pellicle strips around the cell periphery doubles. Each daughter cell (usually) inherits the same number of pellicle strips as the parent cell in a semi-conservative manner. During strip doubling, new strips emerge within the articulation zones between mature strips. In the euglenid clade consisting of photoautotrophic and secondarily osmotrophic lineages, the newly produced pellicle strips do not extend to the posterior tip of the cell and consequently form whorled surface patterns of strip termination (Esson and Leander 2006, 2008; Leander and Farmer 2000a; Leander et al. 2001). Strips that terminate before reaching the posterior tip of the cell occupy a relative position along the length of the cell called a “whorl”. The number of posterior whorls varies between different species, ranging from one to four. In some species, the whorls themselves can be dissociated into one or more subwhorls (Esson and Leander 2008; Leander and Farmer 2000a; Leander et al. 2001). Comparative analyses of the strip termination patterns in several different species have provided important insights into the development processes associated with the control and evolutionary diversification of the euglenid pellicle (Esson and Leander 2006, 2008; Leander and Farmer 2000a; Leander et al. 2001). The complexity of strip surface patterns in some euglenids appears to be either a product of constructive neutrality or a vestige of evolutionary history rather than an adaptation to any particular set of environmental conditions (Esson and Leander 2009, in review).
Evolutionary History of the Euglenid Pellicle
The number of strips around the cell periphery corresponds to the mode of nutrition and locomotion in different euglenid lineages: euglenids that generally eat small prey cells (i.e. bacteriovores) have small rigid cells with less than 12 longitudinal strips; euglenids that generally eat large prey cells (i.e. eukaryovores) are capable of metaboly and have 20 to about 60 strips that are usually helically arranged. Some rigid euglenids with only a few strips (<12) are also capable of eating large prey cells; these euglenids are conspicuously large (>30 microns). Most photoautotrophic (and secondarily osmotrophic) euglenids have a large number of helically arranged strips and are capable of metaboly (Leander et al. 2007). Following the endosymbiotic origin of photosynthesis in one subset of eukaryovorous euglenids, several lineages have independently become rigid with longitudinally arranged strips (see Fig. 7). The number of strips in photoautotrophic euglenids ranges from about 16 to 120. The ancestral state for the number of strips in photoautotrophic euglenids is between 40 and 50; strip numbers that are significantly higher or lower than 40-50 represent derived states. For instance, the cells of some species are relatively enormous and accordingly have the largest known number of strips (e.g. 80 strips in L. helicoideus and 120 strips in E. obtusa) (Esson and Leander 2008; Leander and Farmer 2000b). The photoautotrophic lineages that have lost metaboly, such as Phacus and Lepocinclis (Fig. 7B-E), tend to have 32 strips, which is the inferred ancestral state for the more inclusive clade consisting of these two genera plus Discoplastis. A subgroup of Phacus reduced the number of strips even further to about 20; these cells are among the smallest of all known photoautotrophic euglenids.



Fig. 7. Scanning electron micrographs of photoautotrophic and primary osmotrophic euglenids that have independently lost the ability to undergo metaboly. A. Monomorphina; B-C. Phacus; D-E. Lepocinclis; F. Rhabdomonas. © Brian S. Leander
Primary osmotrophic euglenids diverged from eukaryovorous ancestors independently from the lineage that led to photoautotrophic euglenids. Because primary osmotrophs lost the feeding apparatus in association with this new mode of nutrition, there were concomitant changes in the pellicle. The earliest diverging primary osmotrophs (e.g. Distigma) have retained many of the features found in eukaryovores, such as about 20 helically arranges strips and metaboly; however, many subsequent lineages became rigid, reduced the overall number of strips, and converted the remaining strips to a more longitudinal orientation (e.g. Menoidium and Rhabodmonas) (Fig. 7F) (Leander et al. 2001b). The strips in these lineages are often fused and form a thin and continuous proteinacous layer around the cell.
The evolution of strip number involved at least three mechanisms associated with cytoskeletal replication and cell division: (1) asymmetrical segregation of strips to daughter cells, (2) permanent strip doubling events, and (3) permanent strip halving events (Esson and Leander 2006; Leander 2004; Leander et al. 2001a, 2001b, 2007;). Variation in the number of strips within species indicates that strips are not necessarily distributed evenly during cell division. For instance, a parent cell with 40 strips doubles the number of strips to 80 prior to cell division. In most cases, the two daughter cells will each receive 40 strips and recover the number that was present in the parent cell. In other cases, the daughter cells might receive some other proportion, such as 38 and 42 or 36 and 44. It is also possible that strip duplication is not always faithful; for instance, a parent cell with 40 strips might only produce 39 new strips in which case the daughter cells will receive 39 and 40 strips respectively. Permanent strip duplication events refer to a cell that duplicates its strips but fails to divide. The distribution of strip numbers found in known euglenids suggest that this mechanism happened several times during the evolution of the group; there is evidence for the following events: 4 strip to eight strips; 10 strips to 20 strips; 20 strips to 40 strips (Esson and Leander 2006; Leander et al. 2001a, 2001b, 2007; Leander 2004). Permanent strip halving events refers to a cell that divides without duplicating its strips. This mechanism helps explain the reduction of strips during the evolution of the rigid photoautotrophic lineage Monomorphina (32 strips to 16 strips) (Fig. 7A) (Leander and Farmer 2001b).
To summarize, the euglenid pellicle is very diverse, and comparative analyses have demonstrated a great deal of intermediate states for several cytoskeletal characters. This diversity placed in a molecular phylogenetic context demonstrates many large-scale evolutionary trends within the group (Leander et al. 2007).
Feeding Apparatus
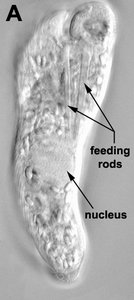
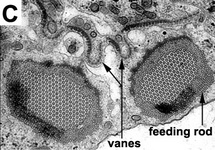
Phagotrophic euglenids have feeding apparatuses that range from relatively simple microtubule reinforced pockets (MtR pockets) to highly sophisticated systems of rods and vanes (Leander et al. 2007; Triemer and Farmer 1991a, 1991b). Bacteriovorous euglenids like Petalomonas and Notosolenus have MtR pockets; the reinforcing microtubules are derived from the ventral root of the flagellar apparatus. However, other bacteriovorous euglenids like Entosiphon and Ploeotia possess feeding apparatuses that are among the most complex of any described so far. These apparatuses consist of two robust rods composed of ordered arrays of microtubules (also derived from the ventral root) embedded within an amorphous matrix (Fig. 8A-D). In Entosiphon, one of the rods bifurcates near the anterior end of the cell and gives the impression of three feeding rods in transverse view (Fig. 8D). The feeding rods in bacteriovorous euglenids and those in some eukaryovorous euglenids, like Dinema, extend the entire length of the cell (Fig. 8B).
By contrast, the feeding rods are reduced in length and are confined to the anterior third of the cell in eukaryovorous euglenids that are capable of rapid metaboly (Fig. 9A), such as Peranema, Urceolus and Jenningsia. A smaller “accessory rod” is usually positioned along the lateral margin of each feeding rod in both bacterivorous and eukaryovorous euglenids (Nisbet 1974). Between the two feeding rods are four to five plicate or lamellar vanes, depending on the species (see Fig. 8B-D). When a prey cell is about to be ingested, the rods of the feeding apparatus protrude from the anterior end of the cell and the vanes twist open like the blades of a pinwheel (Triemer and Fritz 1987). When the feeding apparatus retracts, the vanes twist back into their original position, gripping and internalizing the prey in the process. Although most phagotrophic euglenids ingest their prey whole, some euglenids (e.g. Peranema) can also feed by myzocytosis (Triemer 1997). This mode of feeding is vampire-like, in that the feeding rods pierce the prey cell allowing the cell contents to be sucked into a phagosomal vacuole within the euglenid. The feeding apparatuses present in photoautotrophic and osmotrophic euglenids are highly reduced, corresponding to the switch from phagotrophic modes of nutrition to photosynthesis (via secondary endosymbiosis) and surface absorption, respectively (Shin et al. 2002; Surek and Melkonian 1986; Willey and Wibel 1985).
The ultrastructural diversity of the euglenid feeding apparatus is largely unknown because of the difficulties in reconstructing complex cytoskeletal systems like these using serial TEM sections. Moreover, phagotrophic euglenids are very difficult to cultivate and to isolate from natural samples (i.e. they are not bloom-formers). Much of what is known about the ultrastructure of phagotrophic euglenids has come from micromanipulated samples that have been prepared for transmission electron microscopy one cell at a time.
Chloroplasts
Most euglenids are phagotrophic and lack any evidence of ever having been photoautotrophic in their evolutionary history (Leander 2004). Many euglenids have rigid pellicles and an elaborate feeding apparatus used to capture and ingest bacteria. Eukaryovorous euglenids have dynamic pellicles and routinely consume photoautotrophic microeukaryotes, such as diatoms and single-celled green algae. Photoautotrophic euglenids—or “euglenophytes”—are monophyletic and evolved from eukaryovorous ancestors that established a secondary endosymbiosis with green algal prey cells (Gibbs 1978; Leander 2004). The chloroplasts of euglenophytes are surrounded by three membranes (Fig. 9D) and possess thylakoids in stacks of three. Most euglenid chloroplasts contain a conspicuous pyrenoid (a region containing RuBisCO protein) (see Fig. 9A, C); however, the small disc-shaped chloroplasts of Discoplastis, Lepocinclis and Phacus lack pyrenoids altogether. Crystallized carbohydrate storage products (beta 1,3 glucan), called “paramylon” are often associated with the pyrenoids, but paramylon grains are also distributed throughout the cytoplasm (see Fig. 9A). Chloroplasts with conspicuous paramylon caps on both sides of the pyrenoid are referred to as being “diplopyrenoidal” (Fig. 9).
The number and morphology of chloroplasts within euglenid cells is very diverse (e.g. shield-shaped, disc-shaped and star-shaped) and reflects evolutionary relationships, different stages in cell development and environmental conditions. Some euglenophytes are known to switch nutritional modes and survive in the dark, whereby the chloroplasts become bleached over time. Several different groups of euglenophytes include descendents that have independently lost photosynthesis (e.g. "Khawkinea quartana"—now Euglena quartana, "Astasia longa"—now Euglena longa, "Hyalophacus"—now Phacus and "Cyclidiopsis"—now Lepocinclis); however, highly reduced chloroplasts still exist within these secondary osmotrophs (Hachtel 1998).



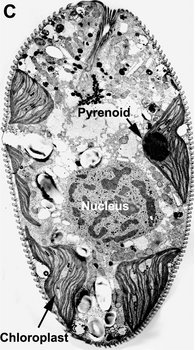

Fig. 9. Micrographs showing the general structure of euglenophyte chloroplasts. A. Light micrograph showing pyrenoids capped with paramylon. © A. Preisfeld & D. J. Patterson. B. Confocal micrograph showing many shield-shaped autofluorescent chloroplasts and muciferous bodies. C. Low magnification transmission electron micrograph showing the peripheral distribution of chloroplasts with pyrenoids. D. High magnification transmission electron micrograph showing three membranes that envelope the chloroplast. B-D © Brian S. Leander, B-C modified from Brown, Leander & Farmer 2002.
Photoreception
Euglenophytes (and most secondary osmotrophs) can respond to the intensity and direction of light and orient themselves in the water column accordingly (Kuznicki et al. 1990). Photoreception is accomplished by an extra-plastidic (i.e. outside of the chloroplast) apparatus consisting of a photosensory swelling at the base of the emergent or swimming flagellum and a closely associated shading structure composed of (orange or red) carotinoids, called the “stigma” or “eyespot” (Fig. 10) The stigma is positioned near the base of an inflated flagellar pocket, often referred to as the “reservoir” in photoautotrophic euglenids. The emergent or swimming flagellum originates from the base of the reservoir and extends through a cylindrical-shaped canal (also a derivative of the flagellar pocket) before exiting the canal opening at the anterior end of the cell. The stigma shades the flagellar swelling only on one side of the cell, so as the cell rotates through the water, the swelling can detect the direction of the most intense light source. For instance, imagine the main light source coming from the sun above. When the cell is oriented with the swelling facing upwards (the stigma oriented down), the swelling will receive a relatively intense flash of light; when the cell is oriented with the swelling facing downwards (the stigma oriented up), the swelling will receive a much less intense flash of light. The behavior of the swimming flagellum will respond in a way that allows the cell to maintain a position in the water column that is optimal for photosynthesis.

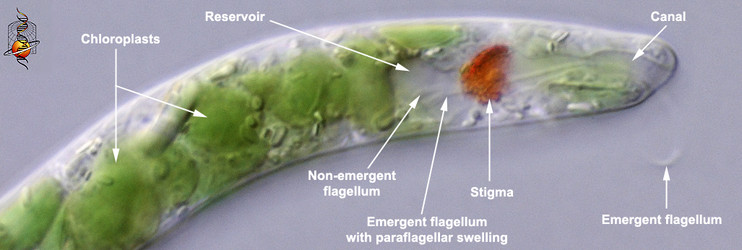
Fig. 10. High magnification light micrograph showing the photoreception apparatus at the anterior end of the cell. © L. Amaral-Zettler and D. J. Patterson.
Interestingly, there is evidence of a photosensory swelling and stigma in a eukaryovorous euglenid called Urceolus (Leander et al. 2001b). No evidence suggests that this organism ever had chloroplasts in its evolutionary history. The photoreception apparatus in Urceolus might enable this predatory euglenid to maintain its position in the photic zone and hunt for photoautotrophic prey. Nonetheless, the evolutionary origin of the photoreception apparatus in Urceolus and photoautotrophic euglenids is unclear; it is clear, however, that the photoreception apparatus in other microeukaryotes (e.g. green algae and dinoflagellates) is almost always embedded within chloroplasts.
Reproduction and Cytokinesis
Sexual reproduction is unknown in euglenids. Asexual reproduction occurs by mitosis followed by cytokinesis. The basal bodies and associated flagellar root system replicate first, followed by the feeding apparatus (if present) and then the pellicle (see Pellicle and Metaboly). After the nucleus and cytoskeleton have duplicated, a cleavage furrow forms at the base of the flagellar pocket near the basal bodies and migrates toward the anterior opening, forming two flagellar pockets within the cell. The cleavage furrow subsequently migrates posteriorly down the longitudinal axis of the cell; the posterior tip of the cell is the last part to become cleaved. The cleavage furrow forms between a (mature) parent strip and a newly generated (nascent) strip on two sides of the cell (Esson and Leander 2006). Each daughter cell (usually) contains the same number of pellicle strips as the parent cell; however, an unequal distribution of strips can also occur during cytokinesis.
Mitochondria
The mitochondria are distinctive in having stalked, paddle-shaped cristae, usually referred to as “discoidal” cristae. Although poorly understood, the mitochondrial genome of euglenids appears to be unusual in having multiple chromosomes (Roy et al. 2007). Transmission electron micrographs indicate that the mitochondria of Petalomonas cantuscygni contain compacted kDNA-like inclusions that are similar in morphology to those found in kinetoplastids (Leander et al. 2001a); however, these inclusions were not noticed in a subsequent study (Roy et al. 2007). Currently, it is unclear whether the mitochondrial genomes of some euglenids contain circular molecules known as ‘maxicircles’ (encoding protein genes) and ‘minicircles’ (encoding ‘guide rRNA’ genes) like those found in kinetoplastids.
Discussion of Phylogenetic Relationships
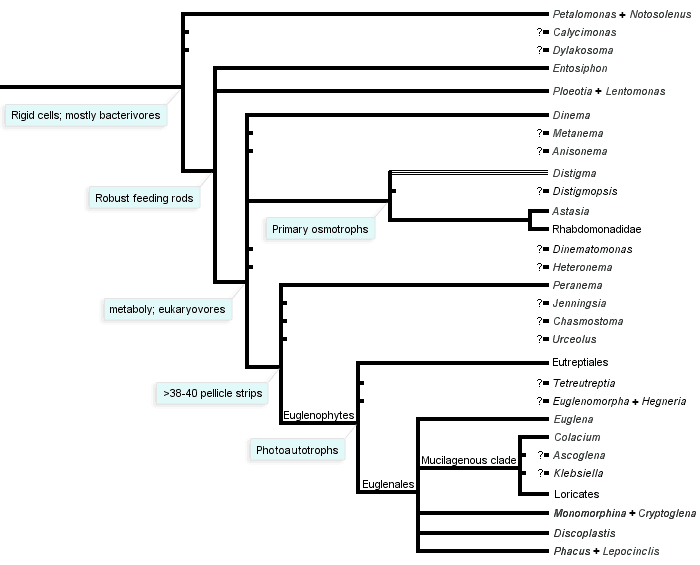
The phylogeny of euglenids has been addressed most extensively using nucleotide sequences amplified from ribosomal genes (i.e. small and large subunit rRNA genes) (Brosnan et al. 2003, 2005; Busse et al. 2003; Linton et al. 1999, 2000; Marin et al. 2003; Milanowski et al. 2006; Montegut-Felkner and Triemer 1997; Preisfeld et al. 2001; Triemer et al. 2006; von der Heyden et al. 2004). Although these genes have been helpful in resolving the phylogeny of photoautotrophic euglenids, they do not provide satisfactory phylogenetic signal at deeper levels in the phylogeny. The most compelling evidence for deep-level phylogenetic relationships of euglenids comes from comparative analyses of morphological data and some nucleus encoded protein genes (e.g. heat shock protein 90) (Breglia et al. 2007; Leander et al. 2001a, 2001b; Montegut-Felkner and Triemer 1997; Simpson et al. 2002; Simpson and Roger 2004; Talke and Preisfeld 2002). These data also indicate that euglenids fall within a putative eukaryotic supergroup called the Excavata, which includes a huge range of free-living and parasitic eukaryotic microbes, such as kinetoplastids (e.g. Bodo and Trypanosoma), diplomonads (e.g. Giardia and Hexamita), parabasalids (e.g. Trichomonas and Trichonympha) and oxymonads (e.g. Saccinobaculus and Streblomastix) (Simpson 2003; Simpson et al. 2006). The nearest sister group to the Euglenida is the clade consisting of diplonemids and kinetoplastids.
Based on the data currently available, we can draw the following conclusions:
- Euglenids are a monophyletic group within the Euglenozoa, and the best synapomorphy for the group is the presence of pellicle strips (fused in some lineages).
- Photoautotrophic euglenids, or euglenophytes, are a monophyletic subgroup nested within a paraphyletic assemblage of phagotrophic lineages (Fig. 11).
- Euglenophytes with one emergent flagellum (Euglenales) are monophyletic; the Eutreptiales, with two emergent flagella, form the nearest sister group.
- Phacus and Lepocinclis are each monophyletic and together form a more inclusive monophyletic group within the euglenophytes; these lineages tend to have 32 pellicle strips, show great diversity in cell shape and possess many small disc-shaped chloroplasts without pyrenoids.
- Discoplastis is monophyletic and possesses morphological features that are similar to Phacus and Lepocinclis (e.g. disc-shaped plastids without pyrenoids and 32 pellicle strips); however, these cells undergo dynamic metaboly, and the molecular phylogenetic position of this lineage within the euglenophytes is currently unresolved.
- The loricates Trachelomonas and Strombomonas are each monophyletic and together form a more inclusive monophyletic group within the euglenophytes.
- The nearest sister group to the loricates is the mucilaginous stalk forming genus Colacium.
- The rigid euglenophytes Monomorphina and Cryptoglena form a monophyletic group; these lineages have a few chloroplasts with stalked pyrenoids and a relatively small number of broad pellicle strips (around 16-20).
- The modern (revised) version of the genus Euglena is monophyletic; these lineages often have 40 pellicle strips, undergo metaboly and show great diversity in cell shape and chloroplast morphology (e.g. shield-shaped, stellate and ribbon-shaped).
- Photosynthesis was lost several times independently within the euglenophytes (e.g. Euglena longa and Euglena quartana—previously “Astasia” and “Khawkinea”, respectively).
- The nearest sister lineages to euglenophytes are eukaryovorous euglenids (e.g. Peranema).
- Primary osmotrophic euglenids (e.g. Distigma, Rhabdomonas and Astasia sensu stricto) are monophyletic and diverged from eukaryovorous ancestors independently from euglenophytes (Fig. 11).
- The single emergent flagellum present in many primary osmotrophs and most euglenophytes evolved independently in these groups in association with an adaptive switch from gliding motility to swimming motility.
- Eukaryovorous euglenids are paraphyletic.
- Bacteriovorous euglenids are paraphyletic.
- Bacteriovorous euglenids, like Petalomonas and Notosolenus, have an unclear molecular phylogenetic position. Comparative morphology indicates that they have retained several ancestral characters, such as few pellicle strips (less than 10), an MtR pocket, kDNA-like mitochondrial inclusions and bacteriovorous modes of nutrition.
- Bacteriovorous euglenids, like Entosiphon and Ploeotia, also have an unclear molecular phylogenetic position. These lineages have pellicles similar to Petalomonas and Notosolenus and complex feeding apparatuses, consisting of rods and vanes, similar to eukaryovorous euglenids.
- Knowledge about the overall diversity and phylogenetic relationships of phagotrophic euglenids (i.e. bacterivores and eukaryovores) is very poor.
Fossil Record
Euglenid fossils are sparse. Aside from the loricas of Trachelomonas and Strombomonas, euglenids do not secrete hard parts that would promote fossilization. However, some photoautotrophic euglenids have exceedingly thick proteinaceous strips, which could presumably fossilize. One enigmatic fossil with euglenid-like features (e.g. strips and a canal opening) was discovered in Silurian deposits, namely Moyeria (Gray 1989). The size, shape and surface morphology of these fossils are reminiscent of some photoautotrophic euglenids in the genus Monomorphina. Apparently, more definitive euglenid fossils have been discovered in rocks from the late Tertiary.
References
Angeler, D. G., A. N. Mullner, and M. Schagerl. 1999. Comparative ultrastructure of the cytoskeleton and nucleus of Distigma (Euglenozoa). European Journal of Protistology 35:309-318.
Belhadri, A., D. Bayle, and G. Brugerolle. 1992. Biochemical and immunological characterization of intermicrotubular cement in the feeding apparatus of phagotrophic eugelnoids: Entosiphon, Peranema, and Ploeotia. Protoplasma 168:113-124.
Belhadri, A., and G. Brugerolle. 1992. Morphogenesis of the feeding apparatus of Entosiphon sulcatum: an immunofluorescence and ultrastructural study. Protoplasma 168:125-135.
Breglia, S. A., C. H. Slamovits, and B. S. Leander. 2007. Phylogeny of phagotrophic euglenids (Euglenozoa) as inferred from hsp90 gene sequences. J. Eukaryot. Microbiol. 52:86-94.
Brosnan, S., P. J. P. Brown, M. A. Farmer, and R. E. Triemer. 2005. Morphological separation of the euglenoid genera Trachelomonas and Strombomonas (Euglenophyta) based on lorica development and posterior strip reduction. J. Phycol. 41:590-605.
Brosnan, S., W. Shin, K. M. Kjer, and R. E. Triemer. 2003. Phylogeny of the photosynthetic euglenophytes inferred from the nuclear SSU and partial LSU rDNA. Int. J. Sys. Evol. Microbiol. 53:1175-1186.
Brown, P. J. P., B. S. Leander, and M. A. Farmer. 2002. Redescription of Euglena rustica (Euglenophyceae), a rare marine euglenophyte from the intertidal zone. Phycologia 41:445-452.
Busse, I., D. J. Patterson, and A. Preisfeld. 2003. Phylogeny of phagotrophic euglenids (Euglenozoa): a molecular appraoch based on culture material and environmental samples. J. Phycol. 39:828-836.
Esson, H. J., and B. S. Leander. 2006. A model for the morphogenesis of strip reduction patterns in phototrophic euglenids: Evidence for heterochrony in pellicle evolution. 8:378-388. Evolution & Development. 8:378-388.
Esson, H. J., and B. S. Leander. 2008. Novel pellicle surface patterns on Euglena obtusa Schmitz (Euglenophyta), a euglenophyte from a benthic marine environment: Implications for pellicle development and evolution. J. Phycol. 43:132-141.
Farmer, M. A., and R. E. Triemer. 1988. Flagellar systems in the euglenoid flagellates. BioSystems 21:283-291.
Gibbs, S. P. 1978. The chloroplasts of Euglena may have evolved from symbiotic green algae. Can. J. Bot. 56
Hachtel, W. 1998. A plastid genome in the heterotrophic flagellate Astasia longa. Endocytobiosis and Cell Research 12:191-193.
Hilenski, L. L., and P. L. Walne. 1985. Ultrastructure of the flagella of the colorless phagotroph Peranema trichophorum (Euglenophyceae. II. Flagellar roots). J. Phycol. 21:125-134.
Kuznicki, L., E. Mikolajczyk, and P. L. Walne. 1990. Photobehavior of euglenoid flagellates: theoretical and evolutionary perspectives. Plant Sci. 9:343-369.
Leander, B. S. 2004. Did trypanosomatid parasites have photosynthetic ancestors? Trends Microbiol . 12:251-258.
Leander, B. S., H. J. Esson, and S. A. Breglia. 2007. Macroevolution of complex cytoskeletal systems in euglenids. BioEssays 29:987-1000.
Leander, B. S., and M. A. Farmer. 2000a. Comparative morphology of the euglenid pellicle. I. Patterns of strips and pores. J. Eukaryot. Microbiol. 47:469-79.
Leander, B. S., and M. A. Farmer. 2000b. Epibiotic bacteria and a novel pattern of strip reduction on the pellicle of Euglena helicoideus (Bernard) Lemmermann. Eur. J. Protistol. 36:405-413.
Leander, B. S., and M. A. Farmer. 2001a. Comparative morphology of the euglenid pellicle. II. Diversity of strip substructure. J. Eukaryot. Microbiol. 48:202-217.
Leander, B. S., and M. A. Farmer. 2001b. Evolution of Phacus (Euglenophyceae) as inferred from pellicle morphology and SSU rDNA. J. Phycol. 37:143-159.
Leander, B. S., R. P. Witek, and M. A. Farmer. 2001a. Trends in the evoluton of the euglenid pellicle. Evolution 55:2115-2135.
Leander, B. S., R. E. Triemer, and M. A. Farmer 2001b. Character evolution in heterotrophic euglenids. Eur. J. Protistol. 37:337-356.
Linton, E. W., D. Hittner, C. Lewandowski, T. Auld, and R. E. Triemer. 1999. A molecular study of euglenoid phylogeny using small subunit rDNA. J Eukaryot Microbiol 46:217-23.
Linton, E. W., M. A. Nudelman, V. Conforti, and R. E. Triemer. 2000. A molecular analysis of the euglenophytes using SSU rDNA. Journal of Phycology 36:740-746.
Marin, B., A. Palm, M. Klingberg, and M. Melkonian. 2003. Phylogeny and taxonomic revision of plastid-containing euglenophytes based on ssu rDNA sequence comparisons and synapomorphic signatures in the ssu rRNA secondary structure. Protist 154:99-145.
Marrs, J. A., and B. Bouck. 1992. The two major membrane skeletal proteins (articulins) of Euglena gracilis define a novel class of cytoskeletal proteins. J. Cell Biol. 118:1465-1475.
Milanowski, R., S. Kosmala, B. Zakrys, and J. Kwiatowski. 2006. Phylogeny of photosynthetic euglenophytes based on combined chloroplast and cytoplasmic SSU rDNA sequence analysis. J. Phycol. 42:721-730.
MontegutFelkner, A. E., and R. E. Triemer. 1997. Phylogenetic relationships of selected euglenoid genera based on morphological and molecular data. Journal of Phycology 33:512-519.
Nisbet, B. 1974. An ultrastructural study of the feeding apparatus of Peranema trichophorum. J. Protozool. 21:39-48.
Preisfeld, A., I. Busse, M. Klingberg, S. Talke, and H. G. Ruppel. 2001. Phylogenetic position and inter-relationships of the osmotrophic euglenids based on SSU rDNA data, with emphasis on the Rhabdomonadales (Euglenozoa). International Journal of Systematic and Evolutionary Microbiology 51:751-758.
Roy, J., D. Faktorova, J. Lukes, and G. Burger. 2007. Unusual mitochondrial genome structures throughout the Euglenozoa. Protist. 158:385-396.
Saito, A., Y. Suetomo, M. Arikawa, G. Omura, S. M. M. K. Khan, S. Kakuta, E. Suzaki, K. Kataoka, and T. Suzaki. 2003. Gliding movement in Peranema trichophorum is powered by flagellar surface motility. Cell Motil. Cytoskel. 55:244-253.
Shin, W., S. M. Boo, and R. E. Triemer. 2001. Ultrastructure of the basal body complex and putative vestigial feeding apparatus in Phacus pleuronectes (Euglenophyceae). Journal of Phycology 37:913-921.
Shin, W., S. Brosnan, and R. E. Triemer. 2002. Are cytoplasmic pockets (MTR/pocket) present in all photosynthetic euglenoid genera? J. Phycol. 38:790-799.
Simpson, A. G. B. 1997. The identity and composition of the Euglenozoa. Archiv Fur Protistenkunde 148:318-328.
Simpson, A. G. B. 2003. Cytoskeletal organisation, phylogenetic affinities and systematics in the contentious taxon Excavata (Eukaryota). Int. J. Sys. Evol. Microbiol. 53:1759-1777.
Simpson, A. G. B., Y. Inagaki, and A. J. Roger. 2006. Comprehensive multi-gene phylogenies of excavate protists reveal the evolutionary positions of ‘primitive’ eukaryotes. Mol. Biol. Evol. 23:615-625.
Simpson, A. G. B., J. Lukes, and A. J. Roger. 2002. The evolutionary history of kinetoplastids and their kinetoplasts. Mol. Biol. Evol. 19:2071-2083.
Simpson, A. G. B., and A. J. Roger. 2004. Protein phylogenies robustly resolve deep-level relationships within Euglenozoa. Mol. Phyl. Evol. 30:201-212.
Solomon, J. A., P. L. Walne, and P. A. Kivic. 1987. Entosiphon sulcatum (Euglenophyceae): flagellar roots of the basal body complex and reservoir regions). J. Phycol. 23:85-98.
Surek, B., and M. Melkonian. 1986. A cryptic cytostome is present in Euglena. Protoplasma 133:39-49.
Talke, S., and A. Preisfeld. 2002. Molecular evolution of euglenozoan paraxonemal rod genes par1 and par2 coincides with phylogenetic reconstruction based on small subunit rDNA data. J. Phycol. 38:995-1003.
Triemer, R. E. 1997. Feeding in Peranema trichophorum revisited (Euglenophyta). Journal of Phycology 33:649-654.
Triemer, R. E., and M. A. Farmer. 1991a. An ultrastructural comparison of the mitotic apparatus, feeding apparatus, flagellar apparatus and cytoskeleton in euglenoids and kinetoplastids. Protoplasma 164:91-104.
Triemer, R. E., and M. A. Farmer. 1991b. The ultrastructural organization of the heterotrophic euglenids and its evolutionary implications. Pp. 185-204 in D. J. Patterson and J. Larsen, eds. The Biology of Free-living Heterotrophic Flagellates. Clarendon Press, Oxford.
Triemer, R. E., and L. Fritz. 1987. Structure and operation of the feeding apparatus in a colorless eugelnoid, Entosiphon sulcatum. J. Protozool. 34:39-47.
Triemer, R. E., E. Linton, W. Shin, A. Nudelman, A. Monfils, M. Bennett, and S. Brosnan. 2006. Phylogeny of the euglenales based upon combined SSU and LSU rDNA sequence comparisons and description of Discoplastis gen. nov (Euglenophyta). J. Phycol. 42:731-740.
von der Heyden, S., E. E. Chao, K. Vickerman, and T. Cavalier-Smith. 2004. Ribosomal RNA phylogeny of bodonid and diplonemid flagelaltes and the evolution of euglenozoa. J. Eukaryot. Microbiol. 51:402-416.
Willey, R. L., and R. G. Wibel. 1985. A cytostome/cytopharynx in green eugelnoid flagellates (Euglenales) and its phylogenetic implications. BioSystems 18:369-376.
Zimba, P. V., M. Rowan, and R. E. Triemer. 2004. Identification of euglenoid algae that produce ichthyotoxin(s). J. Fish Diseases 27:115-117.
Title Illustrations

| Scientific Name | Euglenida |
|---|---|
| Specimen Condition | Dead Specimen |
| Copyright |
© Brian S. Leander

|
About This Page
This page is being developed as part of the Tree of Life Web Project Protist Diversity Workshop, co-sponsored by the Canadian Institute for Advanced Research (CIFAR) program in Integrated Microbial Biodiversity and the Tula Foundation.
Brian S. Leander

The University of British Columbia, Vancouver, British Columbia, Canada
Correspondence regarding this page should be directed to Brian S. Leander at
Page copyright © 2012 Brian S. Leander
 Page: Tree of Life
Euglenida. euglenids or euglenoids.
Authored by
Brian S. Leander.
The TEXT of this page is licensed under the
Creative Commons Attribution-NonCommercial License - Version 3.0. Note that images and other media
featured on this page are each governed by their own license, and they may or may not be available
for reuse. Click on an image or a media link to access the media data window, which provides the
relevant licensing information. For the general terms and conditions of ToL material reuse and
redistribution, please see the Tree of Life Copyright
Policies.
Page: Tree of Life
Euglenida. euglenids or euglenoids.
Authored by
Brian S. Leander.
The TEXT of this page is licensed under the
Creative Commons Attribution-NonCommercial License - Version 3.0. Note that images and other media
featured on this page are each governed by their own license, and they may or may not be available
for reuse. Click on an image or a media link to access the media data window, which provides the
relevant licensing information. For the general terms and conditions of ToL material reuse and
redistribution, please see the Tree of Life Copyright
Policies.
- First online 11 September 2008
- Content changed 10 November 2012
Citing this page:
Leander, Brian S. 2012. Euglenida. euglenids or euglenoids. Version 10 November 2012. http://tolweb.org/Euglenida/97461/2012.11.10 in The Tree of Life Web Project, http://tolweb.org/








 Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site
Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site