Euglenozoa
Brian S. Leander and Alastair Simpson

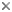
This tree diagram shows the relationships between several groups of organisms.
The root of the current tree connects the organisms featured in this tree to their containing group and the rest of the Tree of Life. The basal branching point in the tree represents the ancestor of the other groups in the tree. This ancestor diversified over time into several descendent subgroups, which are represented as internal nodes and terminal taxa to the right.

You can click on the root to travel down the Tree of Life all the way to the root of all Life, and you can click on the names of descendent subgroups to travel up the Tree of Life all the way to individual species.
For more information on ToL tree formatting, please see Interpreting the Tree or Classification. To learn more about phylogenetic trees, please visit our Phylogenetic Biology pages.
close boxIntroduction
The Euglenozoa is a monophyletic group consisting of single-celled flagellates with very different modes of nutrition, including predation, osmotrophy, parasitism, and photoautotrophy. Predatory euglenozoans are phylogenetically widespread within the group and show a wide diversity of feeding apparatus structure, feeding strategies and prey preferences (Leander et al. 2001; Leander 2004). For instance, some predatory species prefer small prey such as bacteria (e.g., Bodo and Entosiphon); other species, such as Peranema and diplonemids frequently consume larger prey, such as other eukaryotic cells, by either engulfing them whole ( ‘true’ phagotrophy) or by piercing the prey cell and consuming the contents (myzocytosis). Most predatory euglenids are adapted to move and feed on surfaces and they are important components of the microbial biota in many surface sediments. Osmotrophic euglenozoans are heterotrophs that lack a feeding apparatus and absorb nutrients directly from their environments (e.g., Distigma and Rhabdomonas). Photoautotrophy is restricted to a specific subclade of euglenid euglenozoans and originated via secondary endosymbiosis between a eukaryovorous euglenid and a green algal cells (Gibbs 1978; Leander 2004, Leander et al. 2007) (e.g. see the third cell, starting from the left, in the title image). Parasitic (and commensalistic) euglenozoans appear to have evolved independently several times within the group (Simpson et al. 2002, 2006). One subgroup, the trypanosomatids (e.g. see right-hand cell in the title image), include the organisms that cause important human illnesses such as "sleeping sickness", leishmaniases, and Chagas’ disease. Euglenozoans, whether they are parasitic or photoautotrophic, share several derived cytoskeletal features associated with the flagellar apparatus and the feeding apparatus.
Characteristics
- Closed mitosis with an intranuclear spindle
- Paddle-shaped, discoidal mitochondrial cristae
- (Ancestrally with) two flagella: an anteriorly directed dorsal flagellum and a posteriorly directed ventral flagellum
- (Ancestrally with) a flagellar apparatus consisting of three microtubular roots: dorsal root, intermediate root and ventral root.
- Microtubule-reinforced ventral or anterior feeding apparatus (MtR pocket)
- Heteromorphic paraxonemal rods - relatively thick flagella (synapomorphy)
- Tubular extrusomes (synapomorphy)
Discussion of Phylogenetic Relationships
Most euglenozoans are members of two major subgroups: kinetoplastids and euglenids. The best morphological synapomorphy for kinetoplastids is a highly structured mitochondrial inclusion of DNA called a "kinetoplast"; the best morphological synapomorphy for euglenids is a distinctive cortical system of overlapping proteinaceous strips that run longitudinally over the cell surface; this system is called the "euglenid pellicle" (Leander et al. 2007) (e.g. see the third cell, starting from the left, in the title image). The third, much smaller group, diplonemids, are free-living phagotrophs and facultative parasites with sack-shaped cells and short, thin flagella (Kivic and Walne 1984; Kent et al. 1987; Marande et al. 2005; Roy et al. 2007) (e.g. see left-hand cell in the title image). Diplonemid mitochondria contain a few very large flattened cristae, whereas almost all other Euglenozoa have smaller and more numerous discoidal cristae. Most recent molecular phylogenetic analyses support a sister relationship between kinetoplastids and diplonemids (Maslov et al. 1999; Simpson and Roger, 2004; Breglia et al. 2007). Several other euglenozoan lineages, such as Postgaardi and Calkinsia (which both inhabit low-oxygen marine environments, form symbioses with epibiotic bacteria) lack the diagnostic features of the three main subgroups and, therefore, have unclear phylogenetic positions within the Euglenozoa (Simpson et al. 1997; Yubuki et al. In review).
References
Breglia, S.A., Slamovits, C.H. and Leander, B.S. 2007. Phylogeny of phagotrophic euglenids (Euglenozoa) as inferred from hsp90 gene sequences. J. Eukaryot. Microbiol. 52:86-94.
Gibbs, S.P. 1978. The chloroplasts of Euglena may have evolved from symbiotic green algae. Can. J. Bot. 56:2883-2889.
Kent, M.L., Elston, R.A., Neral, T.A. and Sawyer, T.K. 1987. An Isonema-like flagellate (Protozoa, Mastigophora) infection in larval geoduck clams, Panope abrupta. J. Invertebr. Pathol. 50:221-229.
Kivic, P.A. and Walne, P.L. 1984. An evaluation of the possible phylogenetic relationship between Euglenophyta and Kinetoplastida. Orig. Life 13:269-288.
Leander, B.S. 2004. Did trypanosomatid parasites have photosynthetic ancestors? Trends Microbiol. 2004:12:251-258.
Leander, B. S., Triemer, R. E. and Farmer, M. A. 2001. Character evolution in heterotrophic euglenids. Eur. J. Protistol. 37:337-356.
Leander, B.S., Esson, H.J. and Breglia, S.A. 2007. Macroevolution of complex cytoskeletal systems in euglenids. Bioessays 29:987-1000.
Marande, W., Lukes, J. and Burger, G. 2005. Unique mitochondrial genome structure in diplonemids, the sister group of kinetoplastids. Eukaryot. Cell 4:1137-1146.
Maslov, D. A., Yasuhira, S. and Simpson, L. 1999. Phylogenetic affinities of Diplonema within the Euglenozoa as inferred from the SSU rRNA gene and partial COI protein sequences. Protist 150:33-42.
Moreira, D., Lopez-Garcia, P. and Vickerman, K. 2004. An updated view of kinetoplastid phylogeny using environmental sequences and a closer outgroup: proposal for a new classification of the class Kinetoplastea. Int. J. Syst. Evol. Microbiol., 54:1861-1875.
Roy, J., Faktorova, D., Benada, O., Lukes, J. and Burger, G. 2007. Description of Rhynchopus euleeides n. sp. (Diplonemea), a free-living marine euglenozoan. J. Eukaryot. Microbiol. 54:137-145.
Simpson, A.G.B. and Roger, A.J. 2004. Protein phylogenies robustly resolve the deep-level relationships within Euglenozoa. Mol. Phylogenet. Evol. 30:201-212.
Simpson, A.G.B. et al. 2002. The evolutionary history of kinetoplastids and their kinetoplasts. Mol. Biol. Evol. 19:2071-2083.
Simpson, A. G. B., Inagaki, Y. and Roger, A. J. 2006. Comprehensive multigene phylogenies of excavate protists reveal the evolutionary positions of "primitive" eukaryotes. Mol. Biol. Evol., 23:615-625.
Simpson, A.G.B., van den Hoff, J., Bernard, C., Burton, H.R. and Patterson, D.J. 1996/97. The ultrastructure and systematic position of the Euglenozoon Postgaardi mariagerensis, Fehchel et al. Arch Protistenkd, 147:213-225.
Yubuki, N., Edgcomb, V.P., Bernhard, J.M. and Leander, B.S. In review. Cellular identity of a diverse group of deep sea, anoxic euglenozoans with epibiotic bacteria: Ultrastructure and molecular phylogeny of Calkinsia aureus from the Santa Barbara Basin (California, USA). BMC Microbiology.
Title Illustrations

| Scientific Name | Euglenozoa |
|---|---|
| Specimen Condition | Scanning electron micrographs |
| Copyright |
© Brian S. Leander

|
About This Page
This page is being developed as part of the Tree of Life Web Project Protist Diversity Workshop, co-sponsored by the Canadian Institute for Advanced Research (CIFAR) program in Integrated Microbial Biodiversity and the Tula Foundation.
Brian S. Leander

The University of British Columbia, Vancouver, British Columbia, Canada
Alastair Simpson

Dalhousie University, Halifax, Nova Scotia, Canada
Correspondence regarding this page should be directed to Brian S. Leander at and Alastair Simpson at
Page copyright © 2009 Brian S. Leander and Alastair Simpson
 Page: Tree of Life
Euglenozoa.
Authored by
Brian S. Leander and Alastair Simpson.
The TEXT of this page is licensed under the
Creative Commons Attribution-NonCommercial License - Version 3.0. Note that images and other media
featured on this page are each governed by their own license, and they may or may not be available
for reuse. Click on an image or a media link to access the media data window, which provides the
relevant licensing information. For the general terms and conditions of ToL material reuse and
redistribution, please see the Tree of Life Copyright
Policies.
Page: Tree of Life
Euglenozoa.
Authored by
Brian S. Leander and Alastair Simpson.
The TEXT of this page is licensed under the
Creative Commons Attribution-NonCommercial License - Version 3.0. Note that images and other media
featured on this page are each governed by their own license, and they may or may not be available
for reuse. Click on an image or a media link to access the media data window, which provides the
relevant licensing information. For the general terms and conditions of ToL material reuse and
redistribution, please see the Tree of Life Copyright
Policies.
- First online 11 September 2008
- Content changed 11 September 2008
Citing this page:
Leander, Brian S. and Alastair Simpson. 2008. Euglenozoa. Version 11 September 2008 (under construction). http://tolweb.org/Euglenozoa/2405/2008.09.11 in The Tree of Life Web Project, http://tolweb.org/





 Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site
Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site